|
|
|
Find ME
Find ME is a tool to predict LTR mobile elements in not-annotated DNA sequences.
The algorithm used in "Find ME" plugin is rather simple, but provides good results in practice.
Here is a short description of the algorithm:
- Firstly you must provide a HMM profile file built from known conserved domains alignment that
are always found in LTR repeats like reverse transcriptase. The results of HMM search are saved as
annotations for sequence.
You can use HMM Search plugin to build HMM profiles from alignment.
The another tool to create HMM profiles is to use HMMER,
a free Unix command line tool created by Sean Eddy.
- For every signal found in previous step terminal repeats are searched.
Only those repeats that are close to HMM signal and surround it are choosen.
The resulted repeats are also saved as annotation for sequence in a separate group.
You can choose if to search for direct (retrotransposones) or reverse (dna transposones) repeats,
minimal repeats length and distance from HMM signals.
- Now from all of the pairs of HMM signals and terminal repeats, ORF filter selects those
with continuous ORFs between repeats and saves annotations selected into separate group.
There are a set of configurable options how to look for ORFs:
ORF locations, ORF sizes, acceptable stop-codon rate.
Found ORFs are also saved as separate sequence annotations and optionaly as
qualifiers for HMM signals.
- The filtered results are aligned and sorted by similarity. This step is not
implemented in version 1.0 and we use third party programs to accomplish it.
After application is finished you can manually browse and check the results of every step.
Such visual analysis for automaticaly selected results helps to find new interesting
elements and details that are ignored by straighforward algorithm.
The results of every step can be saved as Genbank annotations for sequence and reused latter.
'Find ME' authors:
Vladimir Blinov (ICIG, Novosibirsk, Russia): idea, supervising.
Olga Novikova (ICIG, Novosibirsk, Russia): idea, practical application
Mikhail Fursov (IAE, Novosibirsk, Russia): programming
Screenshots:
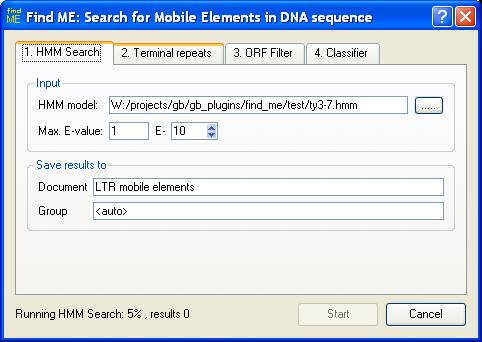
Setting HMM profile, error-level filter for HMM signals and location to store
result annotations.
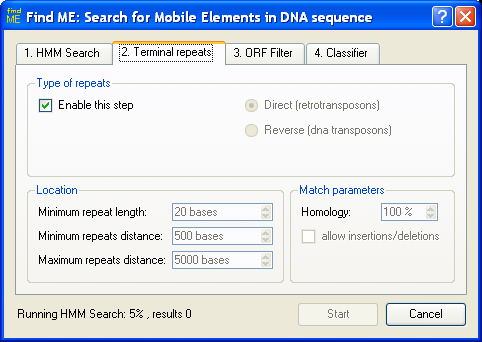
Configuring terminal repeats properties. Note that this step can be disabled.
This can be useful for search of NLRs.
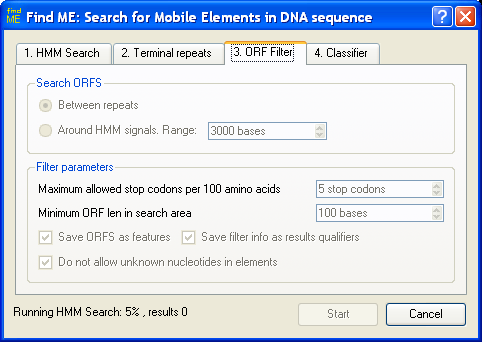
ORF filter configuration.
|
|
|
Febrary, 17, 2006
Find ME 1.0 plugin released.
Search and predict LTR mobile elements in DNA sequences.
|
April, 10, 2005
The site is lauched!
|
|
|

